Research interests
The central goal of our research is to characterize how proteases regulate important biological processes such as apoptosis and cell differentiation. For example, the dysregulation of caspases underlies several human diseases that include cancer and inflammatory disorders, and a better understanding of these enzymes is of great importance in order to design new therapies. Using functional proteomics approaches, our lab focus on characterizing how protease substrates play critical roles in such vital processes, to advances our knowledge about the role of these proteases in human diseases. Harnessing recent progress in cellular engineering, we take advantage of tools such as CRISPR to further support our proteomics findings.

Proteomics in Health and Diseases
Post-Translational Modifications and Protein-Protein Interactions
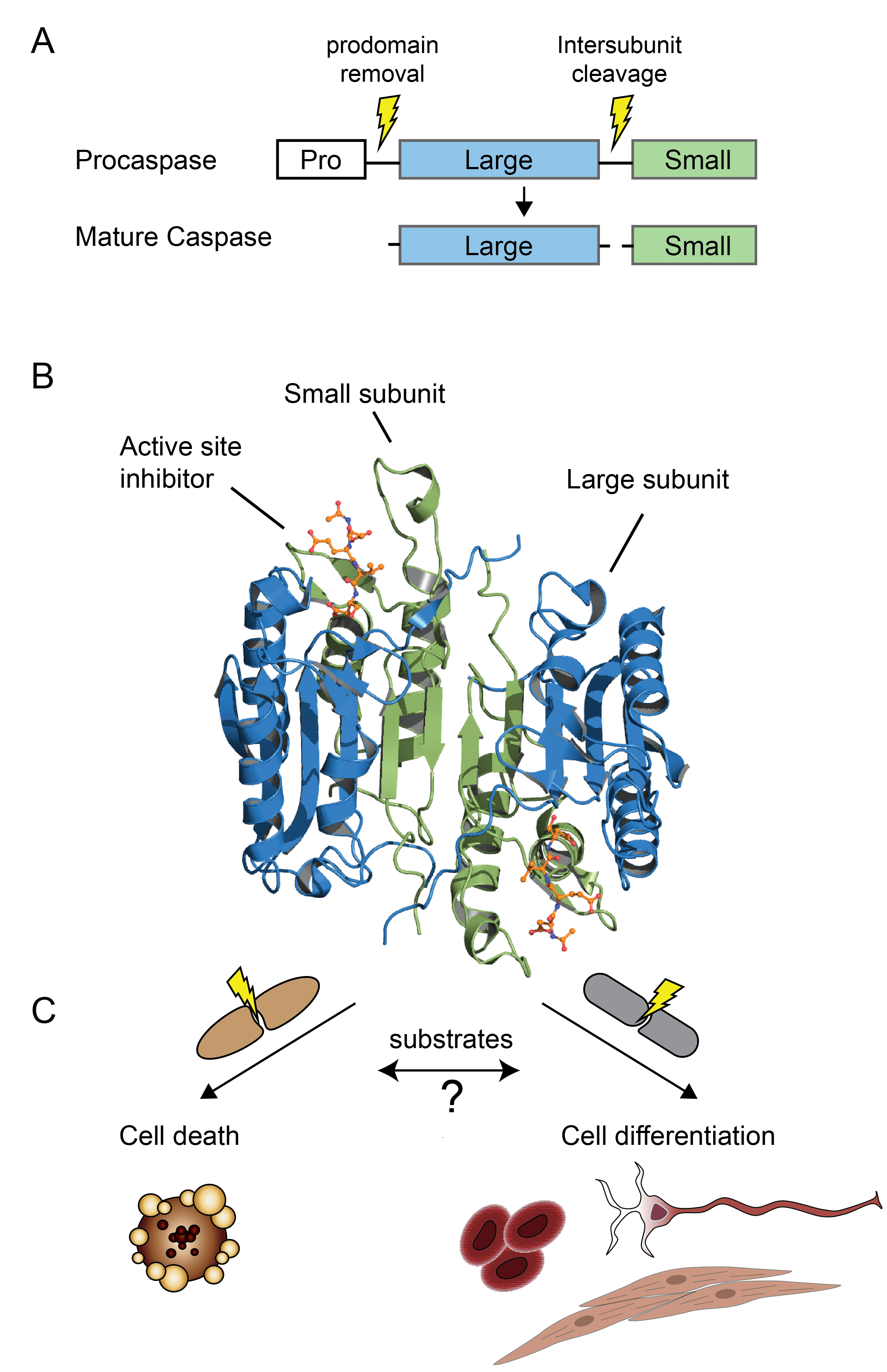
Caspase biology
Caspases are a family of cysteine proteases whose functions are inextricably linked with the process of programmed cell death, or apoptosis, in all metazoans, including C. elegans, Drosophila, mouse and humans. The enzymes predominantly cleave their substrates on the C-terminal side of aspartate. Proteolytic cleavage leads to important changes in cell morphology such as membrane blebbing, DNA fragmentation, phosphatidylserine exposure at the cell surface, and formation of apoptotic vesicles. Recent proteomics approaches allowing the identification of caspase substrates in apoptosis have advanced the field, but our understanding of the role of caspases in other biological processes such as cell differentiation and other programmed forms of cell death like pyroptosis (inflammation) and necroptosis (necrosis) is lagging behind. A deeper understanding of caspase biology relies on many facets, including finding out how each caspase gets activated, what substrates they cleave, what biological functions they each drive, and in what cellular context. One of our research goal is to identify and characterize protease substrates using modern proteomics methods, and how this information can be applied to shed light on the role of caspases and other proteases in biology.
Back to top
Systems biology
Modern proteomic methods using positive enrichment of post-translational modifications can lead to the identification of hundreds or thousands of modified sites on proteins.
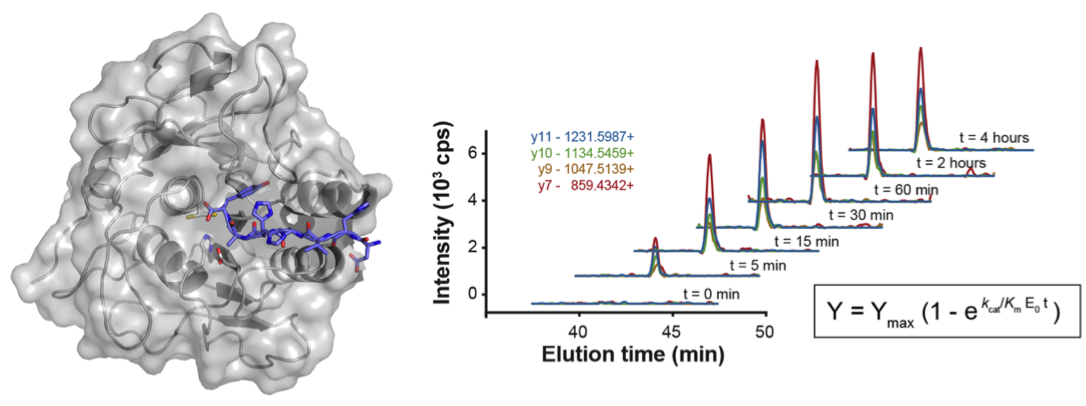
Labeling of protein α-amines from cleavage events provides a powerful approach for profiling proteolysis in complex mixtures since it permits direct identification of protein substrates and the corresponding proteolytic cleavage sites.
One method, developed in the Wells lab, that has proven very powerful in identifying caspase substrates takes advantage of a rationally engineered peptide ligase enzyme called subtiligase to attach ester peptide probes to free N-termini (Mahrus et al., 2008).
In short, the subtiligase-based N-terminomics methodology allows the attachment of a biotin tag onto the free α-amine of the cleaved N-terminal of protein fragments.
The biotin labeled proteins are bound to avidin beads and trypsinized.
The N-terminal peptide is released by cleaving the unique TEV protease cleavage sequence in the tag followed by identification by LC-MS/MS.
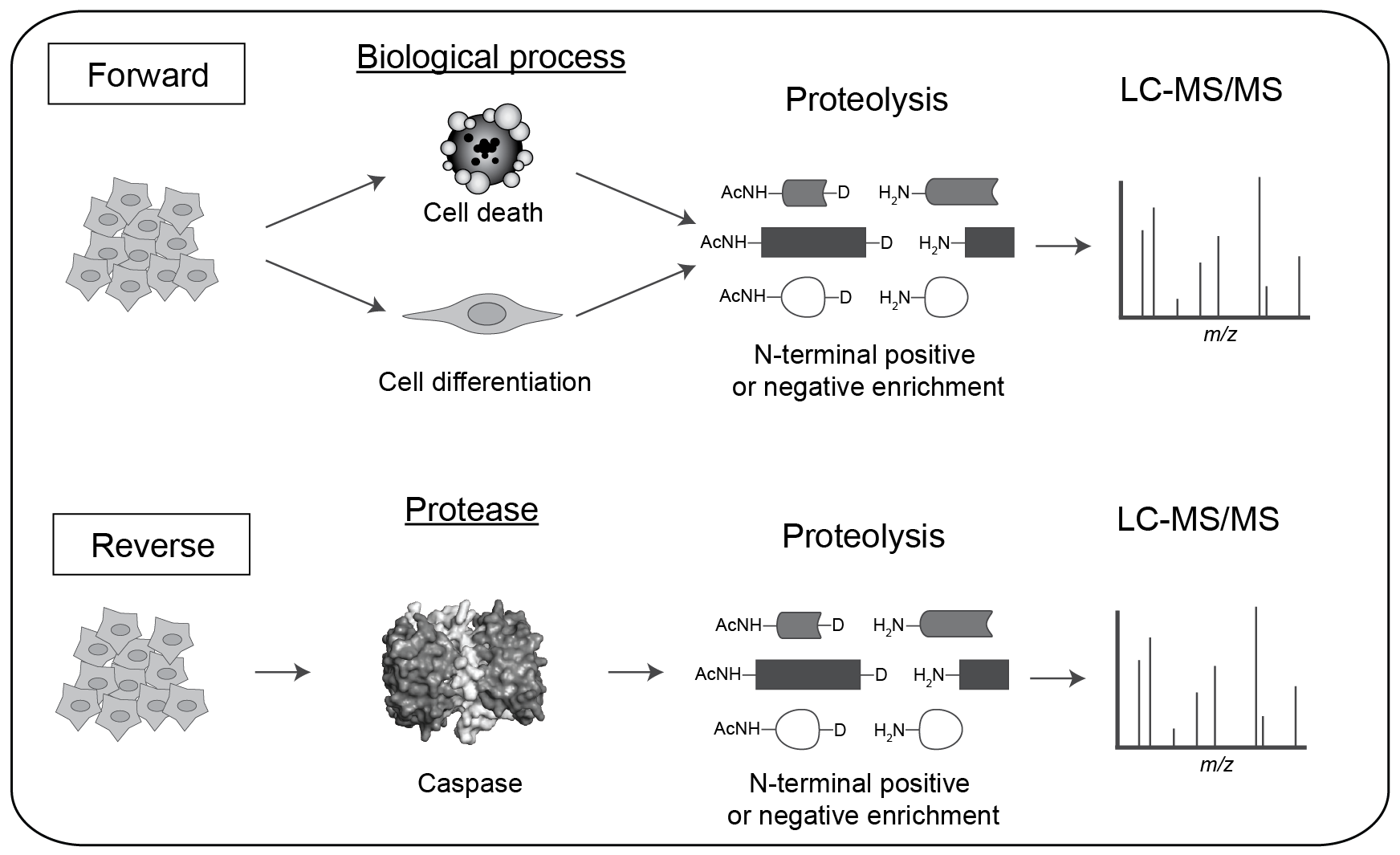
One can use this method to identify native protein substrates either in intact cells by triggering protease activation (the Forward approach) or in cell lysates by addition of exogenous protease (the Reverse approach).
The advantage of the Forward experiment is that one can identify substrates in intact cellular structures with endogenous caspases for the biological process studied, such as cell death or cell differentiation.
However, one cannot easily determine the specific caspase(s) responsible for the proteolytic events.
The Reverse experiment, on the other hand, enables identification of potential substrates after addition of a specific caspase to a cell lysate (neutralized of endogenous proteases).
This is a good method to link proteases to their protein substrates, but does so in an extract where cell architecture and cellular organelles have been destroyed, and requires controls that ensure cleavage events are not the result of indirect activation of endogenous proteases.
Forward and Reverse approaches are very complementary and each helps to validate the other in terms of biological relevance and identity of the specific caspase(s) responsible.
Back to top
Post-Translational Modifications (PTMs) and Protein-Protein Interactions (PPI)


(see our latest publications for more details)
PTMs
- Proteolysis
- Ubiquitination
- Phosphorylation
PPI
- TurboID
- APEX
- Biotinylation by antibody recognition (BAR)
- Immunoprecipitation and mass spectrometry (IP-MS)
Diseases
- Infectious diseases
- Multiple sclerosis
- Cancer
- Cardiovascular diseases